How do we deploy the virtual cluster for the course "Scientific Computing Essentials"?
The Scientific Computing Essentials is the first ever hands-on scientific computing online course that supports playgrounds loaded with the High Performance Computing (HPC) systems software stack: Slurm, PBS Pro, OpenMP, MPI and CUDA!
For this course, we have created a small private virtual cluster with Docker technology. It has the following components:
slurmctld: master node/ login nodemysqlandslurmdbd: database nodesc1: compute node 1c2: compute node 2
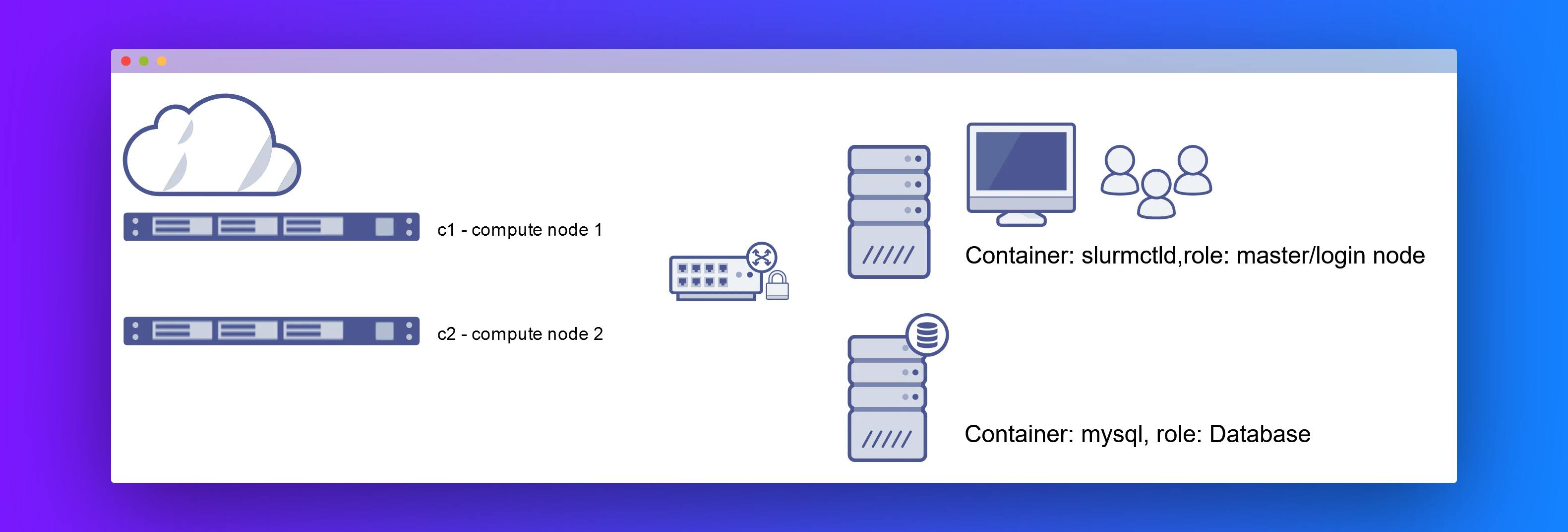
Slurm setup
We created a multi-container Slurm cluster using docker-compose on a Digitial Ocean droplet (ideally we would use multi-droplets). The compose file creates named volumes for persistent storage of MySQL data files as well as Slurm state and log directories.
You can also get the Docker composer files from the https://github.com/giovtorres/slurm-docker-cluster.
Build the image locally:
docker build -t slurm-docker-cluster:19.05.1 .
Register the Cluster with SlurmDBD
docker exec slurmctld bash -c "/usr/bin/sacctmgr --immediate add cluster name=linux" && \
docker-compose restart slurmdbd slurmctld
Accessing the Cluster
We use docker exec to run a bash shell on the controller container:
docker exec -it slurmctld bash
From the shell, we can execute slurm commands, for example:
[root@slurmctld /]# sinfo
PARTITION AVAIL TIMELIMIT NODES STATE NODELIST
normal* up 5-00:00:00 2 idle c[1-2]
Submitting Jobs
The slurm_jobdir named volume is mounted on each Slurm container as /data. Therefore, in order to see job output files while on the controller, change to the /data directory when on the slurmctld container and then submit a job:
[root@slurmctld /]# cd /data/
[root@slurmctld data]# sbatch --wrap="uptime"
Submitted batch job 2
[root@slurmctld data]# ls
slurm-2.out
Stopping and Restarting the Cluster
docker-compose stop
docker-compose start
Deleting the Cluster
To remove all containers and volumes, we run:
docker-compose stop
docker-compose rm -f
docker volume rm slurm-docker-cluster_etc_munge slurm-docker-cluster_etc_slurm slurm-docker-cluster_slurm_jobdir slurm-docker-cluster_var_lib_mysql slurm-docker-cluster_var_log_slurm
PBS Pro setup
To setup PBS Pro, we follow the guideline from the PBS Pro at
Using Docker to Instantiate PBS. However, we first install the PBS Pro on the slurmctld Docker container. Start it
sudo /etc/init.d/pbs start
Before we can submit and run jobs, we add some configurations using root account. Exit the current shell and you should return to a root shell. Run:
qmgr -c "create node pbs"
qmgr -c "set node pbs queue=workq"
This will create a node named pbs and add a queue to it.
Submit a PBS job
To submit and view jobs.
qsub -- /bin/sleep 10
qstat
MPI setup
We setup MVAPICH and OpenMPI on the slurmctld that is setup to use the nodes c1 and c2 as the compute nodes and use \data as the shared folder.
Integration
Finally, we use a Docker terminal redirection technology (TTYD, to be discussed later) to forward the terminals to out the Scientific Computing Essentials course.
PBS Pro job submission

Slurm job submission

This is how create a simple interface to demonstrate PBS Pro and Slurm with a Jupyter notebook like simple user experience! and the best of it, the course is offered for FREE!
Get it now!
Enroll at the Scientific Computing Essentials, Scientific Computing School.